-Search query
-Search result
Showing 1 - 50 of 71 items for (author: park & kh)

EMDB-43033: 
Structure of 80alpha portal protein expressed in E. coli
Method: single particle / : Kizziah JL, Mukherjee A, Dokland T

EMDB-43142: 
SaPI1 mature capsid structure containing DNA
Method: single particle / : Mukherjee A, Kizziah JL, Dokland T

EMDB-43143: 
SaPI1 mature capsid structure without DNA
Method: single particle / : Mukherjee A, Kizziah JL, Dokland T

EMDB-43145: 
SaPI1 portal structure in mature capsids containing DNA
Method: single particle / : Kizziah JL, Mukherjee A, Dokland T

EMDB-43146: 
SaPI1 portal structure in mature capsids without DNA
Method: single particle / : Mukherjee A, Kizziah JL, Dokland T

EMDB-43147: 
SaPI1 portal-capsid interface in mature capsids with DNA
Method: single particle / : Kizziah JL, Mukherjee A, Dokland T

PDB-8v8b: 
Structure of 80alpha portal protein expressed in E. coli
Method: single particle / : Kizziah JL, Mukherjee A, Dokland T

PDB-8vd4: 
SaPI1 mature capsid structure containing DNA
Method: single particle / : Mukherjee A, Kizziah JL, Dokland T

PDB-8vd5: 
SaPI1 mature capsid structure without DNA
Method: single particle / : Mukherjee A, Kizziah JL, Dokland T

PDB-8vd8: 
SaPI1 portal structure in mature capsids containing DNA
Method: single particle / : Kizziah JL, Mukherjee A, Dokland T

PDB-8vdc: 
SaPI1 portal structure in mature capsids without DNA
Method: single particle / : Mukherjee A, Kizziah JL, Dokland T

PDB-8vde: 
SaPI1 portal-capsid interface in mature capsids with DNA
Method: single particle / : Kizziah JL, Mukherjee A, Dokland T

PDB-8c5v: 
Chemotaxis core signalling unit from E protein lysed E. coli cells
Method: subtomogram averaging / : Cassidy CK, Qin Z, Zhang P

EMDB-27703: 
Structure of RBD directed antibody DH1047 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interace
Method: single particle / : May AJ, Manne K, Acharya P

PDB-8dtk: 
Structure of RBD directed antibody DH1047 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interace
Method: single particle / : May AJ, Manne K, Acharya P

EMDB-15641: 
Structure of the Native Chemotaxis Core Signalling Complex from E-gene lysed E. coli cells.
Method: subtomogram averaging / : Qin Z, Zhang P

EMDB-15642: 
Native Chemotaxis Core Signalling Complex from E. coli, with focused alignment on the baseplate (CheA-CheW)
Method: subtomogram averaging / : Qin Z, Zhang P

EMDB-15643: 
Native Chemotaxis Core Signalling Complex from E. coli, Focused alignment on ligand binding domain
Method: subtomogram averaging / : Qin Z, Zhang P
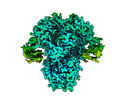
EMDB-26727: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)
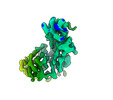
EMDB-26729: 
CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0)
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)
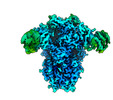
EMDB-26730: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)
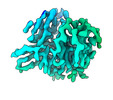
EMDB-26731: 
CCoV-HuPn-2018 S in the swung out conformation (local refinement of domain 0)
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-7us6: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-7us9: 
CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0)
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-7usa: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-7usb: 
CCoV-HuPn-2018 S in the swung out conformation (local refinement of domain 0)
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)
EMDB-26676: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-26677: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains)
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-26678: 
An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-27043: 
Negative stain electron microscopy single particle reconstruction of monoclonal antibody SP1-77 Fab in complex with SARS-CoV2 2P spike
Method: single particle / : Edwards RJ, Mansouri K
EMDB-27044: 
Negative stain electron microscopy single particle reconstruction of monoclonal antibody VHH7-7-53 Fab in complex with SARS-CoV2 2P spike
Method: single particle / : Edwards RJ, Mansouri K

EMDB-27046: 
Negative stain electron microscopy single particle reconstruction of monoclonal antibody VHH7-5-82 Fab in complex with SARS-CoV2 2P spike
Method: single particle / : Edwards RJ, Mansouri K

PDB-7upw: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

PDB-7upx: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains)
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

PDB-7upy: 
An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-25105: 
Structure of SARS-CoV S protein in complex with Receptor Binding Domain antibody DH1047
Method: single particle / : Gobeil S, Acharya P

PDB-7sg4: 
Structure of SARS-CoV S protein in complex with Receptor Binding Domain antibody DH1047
Method: single particle / : Gobeil S, Acharya P

EMDB-21514: 
map of the mammalian Mediator complex
Method: single particle / : Zhao H, Young N, Asturias F

PDB-6w1s: 
Atomic model of the mammalian Mediator complex
Method: single particle / : Zhao H, Young N, Asturias F

EMDB-22309: 
Dimeric Immunoglobin A (dIgA)
Method: single particle / : Kumar Bharathkar S, Parker BP, Malyutin AG, Stadtmueller BM

EMDB-22310: 
Secretory Immunoglobin A (SIgA)
Method: single particle / : Kumar Bharathkar S, Parker BP, Malyutin AG, Stadtmueller BM

PDB-7jg1: 
Dimeric Immunoglobin A (dIgA)
Method: single particle / : Kumar Bharathkar S, Parker BP, Malyutin AG, Stadtmueller BM

PDB-7jg2: 
Secretory Immunoglobin A (SIgA)
Method: single particle / : Kumar Bharathkar S, Parker BP, Malyutin AG, Stadtmueller BM

EMDB-22829: 
Human Tom70 in complex with SARS CoV2 Orf9b
Method: single particle / : QCRG Structural Biology Consortium

PDB-7kdt: 
Human Tom70 in complex with SARS CoV2 Orf9b
Method: single particle / : QCRG Structural Biology Consortium

PDB-6zet: 
Crystal structure of proteinase K nanocrystals by electron diffraction with a 20 micrometre C2 condenser aperture
Method: electron crystallography / : Evans G, Zhang P, Beale EV, Waterman DG

PDB-6zeu: 
Crystal structure of proteinase K lamella by electron diffraction with a 50 micrometre C2 condenser aperture
Method: electron crystallography / : Evans G, Zhang P, Beale EV, Waterman DG

PDB-6zev: 
Crystal structure of proteinase K lamellae by electron diffraction with a 20 micrometre C2 condenser aperture
Method: electron crystallography / : Evans G, Zhang P, Beale EV, Waterman DG

EMDB-20391: 
CryoEM structure of mouse mediator subunit one deletion
Method: single particle / : Asturias F, Zhao H, Young N

EMDB-20392: 
CryoEM structure of wild type mouse mediator with MED19-FLAG
Method: single particle / : Asturias F, Zhao H
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
